EVD transmission model from Khan et al 2015 using Python/PyGOM
Author: Thomas Finnie @twomagpi
Date: 2018-10-03
# housekeeping
import numpy
from pygom import DeterministicOde, TransitionType, SimulateOde, Transition
# first set up the states
states = ['Susceptible_lr',
'Susceptible_hr',
'Exposed',
'Infected',
'Hospitalized',
'Recovered'
]
Table 1
| Parameter | Description | Value |
|---|---|---|
| $\phi_H$ | Modification parameter for infection rate of high-risk susceptible individuals | 1.2-2 |
| $\sigma_I$ | Disease-induced death rate of infected individuals | 0.10 |
| $\sigma_H$ | Disease-induced death rate of hospitalized individuals | 0.5 |
| $\theta_I$ | Recovery rate of infected individuals | 0.1 |
| $\theta_H$ | Recovery rate of hospitalized individuals | 0.2 |
| $\alpha$ | Rate at which latent individuals become infectious | 0.1 |
| $\tau$ | Hospitalization rate for infected individuals | 0.16 |
| $\Pi$ | Recruitment rate | 1.7 |
| p | Fraction of the individuals at high-risk | 0.2 |
| $\beta$ | Transmission rate of disease | Estimated |
| $1/\mu$ | Average life of human | 63 years |
# now set up the parameters
parameters = ['phi_H',
'sigma_I',
'sigma_H',
'theta_I',
'theta_H',
'alpha',
'tau',
'Rec_rate',
'p',
'beta',
'mu',
'lda',
'eta',
'N'
]
# Set this up as birth, death and transitions
# This could have been done as lifts of the ODE system from the paper but
# being more verbose like this makes us concentrate on the model
transitions = [
Transition(origin='Susceptible_lr',
destination='Exposed',
equation='lda*Susceptible_lr',
transition_type=TransitionType.T),
Transition(origin='Susceptible_hr',
destination='Exposed',
equation='phi_H * lda * Susceptible_hr',
transition_type=TransitionType.T),
Transition(origin='Exposed',
destination='Infected',
equation='alpha * Exposed',
transition_type=TransitionType.T),
Transition(origin='Infected',
destination='Hospitalized',
equation='tau * Infected',
transition_type=TransitionType.T),
Transition(origin='Infected',
destination='Recovered',
equation='theta_I * Infected',
transition_type=TransitionType.T),
Transition(origin='Hospitalized',
destination='Recovered',
equation='theta_H * Infected',
transition_type=TransitionType.T),
]
births_deaths = [
# Births first
Transition(origin='Susceptible_lr',
equation='Rec_rate * (1 - p)',
transition_type=TransitionType.B),
Transition(origin='Susceptible_hr',
equation='Rec_rate * p',
transition_type=TransitionType.B),
# Natural Deaths
Transition(origin='Susceptible_lr',
equation='mu * Susceptible_lr',
transition_type=TransitionType.D),
Transition(origin='Susceptible_hr',
equation='mu * Susceptible_hr',
transition_type=TransitionType.D),
Transition(origin='Exposed',
equation='mu * Exposed',
transition_type=TransitionType.D),
Transition(origin='Infected',
equation='mu * Infected',
transition_type=TransitionType.D),
Transition(origin='Hospitalized',
equation='mu * Hospitalized',
transition_type=TransitionType.D),
Transition(origin='Recovered',
equation='mu * Recovered',
transition_type=TransitionType.D),
#disease deaths
Transition(origin='Infected',
equation='sigma_I * Infected',
transition_type=TransitionType.D),
Transition(origin='Hospitalized',
equation='sigma_H * Hospitalized',
transition_type=TransitionType.D)
]
derived_parameters = [('lda', 'beta * ((Infected + (eta*Hospitalized))/N)'),
('N', 'Susceptible_lr + Susceptible_hr + Exposed + Infected + Hospitalized + Recovered')]
# build the model system
model_system = DeterministicOde(states,
parameters,
transition=transitions,
birth_death=births_deaths,
derived_param=derived_parameters
)
model_system.print_ode()
⎡ -(Susceptibleₗᵣ⋅β⋅(Hospitalized⋅η + Infecte
⎢dSusceptible_lr/dt= ───────────────────────────────────────────
⎢
⎢
⎢ -Susceptibleₕᵣ⋅β⋅φ_H⋅(Hospitalized⋅η + I
⎢dSusceptible_hr/dt= ────────────────────────────────────────
⎢
⎢
⎢ -Exposed⋅(α + μ)⋅(Exposed + Hospitalized + Infected + Re
⎢ dExposed/dt= ────────────────────────────────────────────────────────
⎢
⎢
⎢ dInfected/dt=
⎢
⎢ dHospitalized/dt=
⎢
⎣ dRecovered/dt=
d) + (Recᵣₐₜₑ⋅(p - 1) + Susceptibleₗᵣ⋅μ)⋅(Exposed + Hospitalized + Infected +
──────────────────────────────────────────────────────────────────────────────
Exposed + Hospitalized + Infected + Recovered + Susceptibleₕᵣ + Susceptibleₗᵣ
nfected) + (Recᵣₐₜₑ⋅p - Susceptibleₕᵣ⋅μ)⋅(Exposed + Hospitalized + Infected +
──────────────────────────────────────────────────────────────────────────────
Exposed + Hospitalized + Infected + Recovered + Susceptibleₕᵣ + Susceptibleₗᵣ
covered + Susceptibleₕᵣ + Susceptibleₗᵣ) + Susceptibleₕᵣ⋅β⋅φ_H⋅(Hospitalized⋅η
──────────────────────────────────────────────────────────────────────────────
Exposed + Hospitalized + Infected + Recovered + Susceptibleₕᵣ + Susceptibleₗᵣ
Exposed⋅α - Infected⋅μ - Infected⋅σ_I - Infected⋅τ - Infected⋅θ_I
-Hospitalized⋅μ - Hospitalized⋅σ_H + Infected⋅τ - Infected⋅θ_H
Infected⋅θ_H + Infected⋅θ_I - Recovered⋅μ
Recovered + Susceptibleₕᵣ + Susceptibleₗᵣ)) ⎤
──────────────────────────────────────────── ⎥
⎥
⎥
Recovered + Susceptibleₕᵣ + Susceptibleₗᵣ) ⎥
────────────────────────────────────────── ⎥
⎥
⎥
+ Infected) + Susceptibleₗᵣ⋅β⋅(Hospitalized⋅η + Infected)⎥
──────────────────────────────────────────────────────────⎥
⎥
⎥
⎥
⎥
⎥
⎥
⎦
# Create the timeline
t = numpy.linspace(0, 200, 200)
# Set the inital values for the states
model_system.initial_values = ([1000000,
20000,
15,
10,
0,
0],
t[0])
liberia_parameters = {
'phi_H': 1.6,
'sigma_I': 0.1,
'sigma_H': 0.5,
'theta_I': 0.1,
'theta_H': 0.2,
'alpha': 0.1,
'tau': 0.16,
'Rec_rate': 1.7,
'p': 0.2,
'beta': 0.371,
'mu': (1/63)/365,
'eta': 0.7,
}
sierra_leone_parameters = {
'phi_H': 1.6,
'sigma_I': 0.1,
'sigma_H': 0.5,
'theta_I': 0.1,
'theta_H': 0.2,
'alpha': 0.1,
'tau': 0.16,
'Rec_rate': 1.7,
'p': 0.2,
'beta': 0.371,
'mu': (1/63)/365,
'eta': 0.7,
}
model_system.parameters = liberia_parameters
solution,output = model_system.integrate(t[1::], full_output=True)
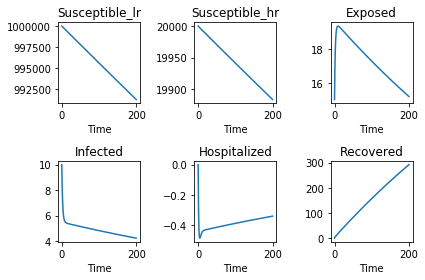
model_system.plot()
<Figure size 640x480 with 6 Axes>
model_system.parameters = sierra_leone_parameters
solution,output = model_system.integrate(t[1::], full_output=True)
model_system.plot()