Stochastic SIR model using odin
Author: Thibaut Jombart @thibautjombart
Date: 2018-10-03
Requirements
This code uses odin, an R package for describing an solving differential equations. The commented commands will install the latest version of the package and its dependencies:
#if (!require("drat")) install.packages("drat")
#drat:::add("mrc-ide")
#install.packages("dde")
#install.packages("odin")
library(odin)
Basic model
odin models can be specified in a separate source file, or directly in an R script, as below. Note that while odin code resembles R, it is not R code code per se - not everything that works in R may work in odin code.
The discrete, stochastic SIR model can be formulated in odin as follows:
sir_generator <- odin::odin({
## Core equations for transitions between compartments:
update(S) <- S - n_SI
update(I) <- I + n_SI - n_IR
update(R) <- R + n_IR
## Individual probabilities of transition:
p_SI <- 1 - exp(-beta * I / N) # S to I
p_IR <- 1 - exp(-gamma) # I to R
## Draws from binomial distributions for numbers changing between
## compartments:
n_SI <- rbinom(S, p_SI)
n_IR <- rbinom(I, p_IR)
## Total population size
N <- S + I + R
## Initial states:
initial(S) <- S_ini
initial(I) <- I_ini
initial(R) <- 0
## User defined parameters - default in parentheses:
S_ini <- user(1000)
I_ini <- user(1)
beta <- user(0.2)
gamma <- user(0.1)
}, verbose = FALSE)
The model is first parsed and compiled using odin::odin, and user-provided parameters are passed using the resulting model generator (the object sir_generator):
sir <- sir_generator(I_ini = 10)
sir
<odin_model>
Public:
clone: function (deep = FALSE)
contents: function ()
graph_data: function ()
init: 1000 10 0
initial: function (step)
initialize: function (user = NULL)
name: odin
names: step S I R
ptr: externalptr
run: function (step, y = NULL, ..., use_names = TRUE, replicate = NULL)
set_user: function (..., user = list(...))
transform_variables: function (y)
update: function (step, y)
update_cache: function ()
user_info: function ()
variable_order: list
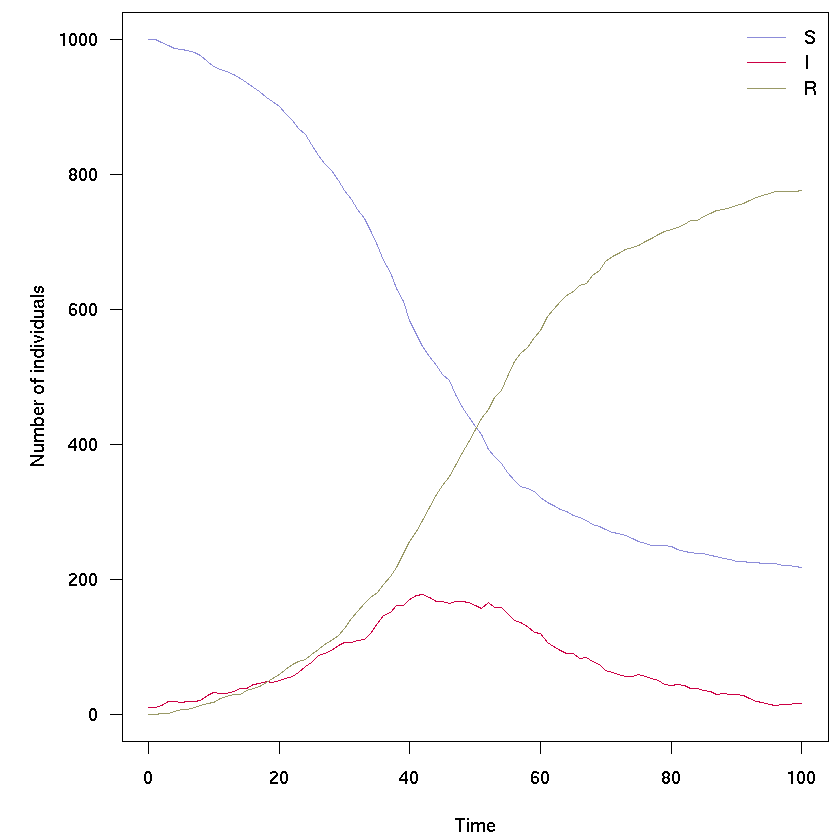
set.seed(1)
sir_col <- c("#8c8cd9", "#cc0044", "#999966")
res <- sir$run(0:100)
par(mar = c(4.1, 5.1, 0.5, 0.5), las = 1)
matplot(res[, 1], res[, -1], xlab = "Time", ylab = "Number of individuals",
type = "l", col = sir_col, lty = 1)
legend("topright", lwd = 1, col = sir_col, legend = c("S", "I", "R"), bty = "n")
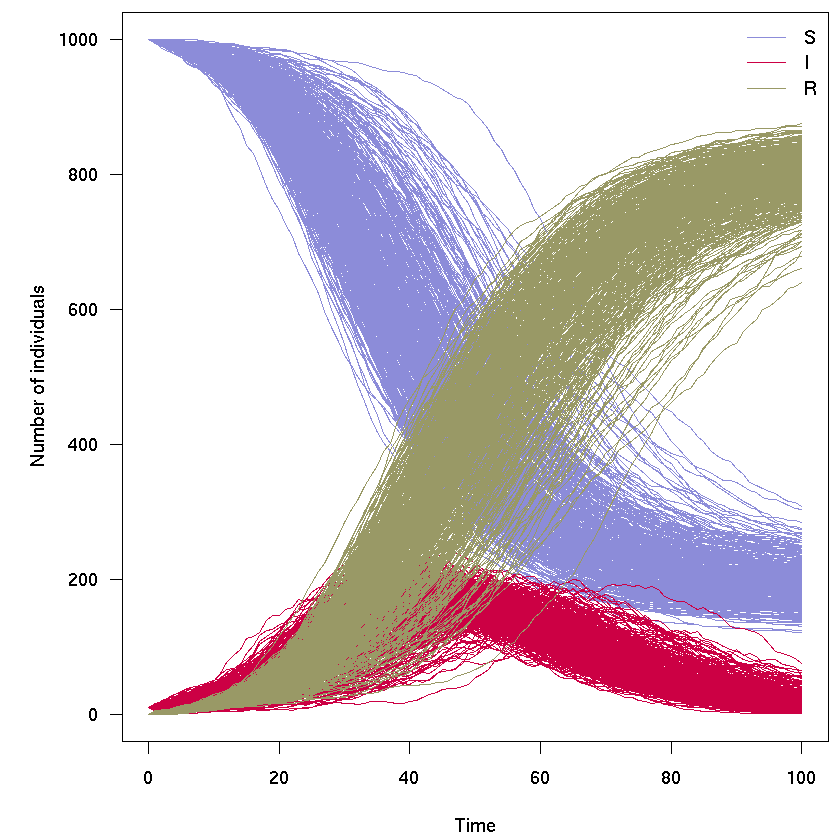
Running multiple simulations
In the following we illustrate how the odin model sir can be used to generate replicates, i.e. several independent runs of the model; it takes a bit of reformatting the output - see next section for some helpful wrappers:
res_200 <- sir$run(0:100, replicate = 200)
res_200 <- sir$transform_variables(res_200)
res_200 <- cbind.data.frame(t = res_200[[1]], res_200[-1])
col <- rep(sir_col, each = 200)
par(mar = c(4.1, 5.1, 0.5, 0.5), las = 1)
matplot(res_200[, 1], res_200[, -1], xlab = "Time", ylab = "Number of individuals",
type = "l", col = col, lty = 1)
legend("topright", lwd = 1, col = sir_col, legend = c("S", "I", "R"), bty = "n")
Simplified workflow
Here we provide some helper functions for formatting odin output with multiple replicates, and plotting the results
Helper functions
## x: instance of odin model
## t: time steps
## n: number of replicates
run_model <- function(x, t = 0:100, n = 1, ...) {
res <- x$run(t, replicate = n, ...)
res <- x$transform_variables(res)
res <- cbind.data.frame(t = res[[1]], res[-1])
attr(res, "n_compartments") <- length(x$names) - 1
attr(res, "n_replicates") <- n
attr(res, "compartments") <- x$names[-1]
class(res) <- c("pretty_odin", class(res))
res
}
sir_pal <- colorRampPalette(sir_col)
plot.pretty_odin <- function(x, pal = sir_pal, ...) {
## handle colors
n_compartments <- attr(x, "n_compartments")
n_replicates <- attr(x, "n_replicates")
col_leg <- pal(n_compartments)
alpha <- max(10 / n_replicates, 0.05)
col <- rep(col_leg, each = n_replicates)
## make plot
par(mar = c(4.1, 5.1, 0.5, 0.5), las = 1)
matplot(x[, 1], x[, -1], xlab = "Time", ylab = "Number of individuals",
type = "l", col = col, lty = 1, ...)
legend("topright", lwd = 1, col = col_leg, bty = "n",
legend = attr(x, "compartments"))
}
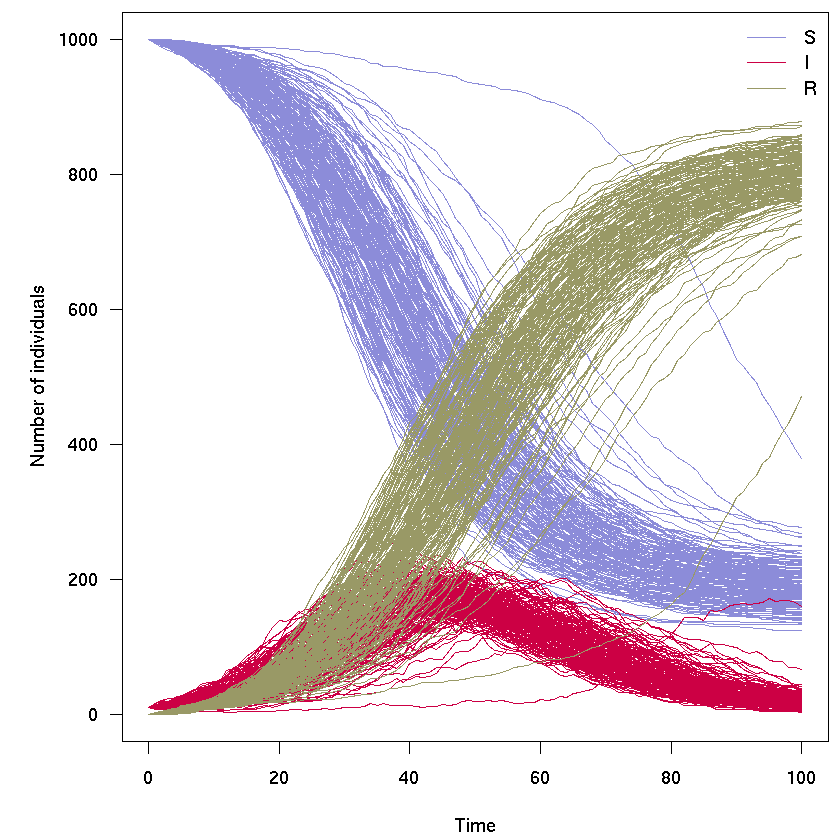
Example of simulations
With 120 time steps, 500 independent replicates:
x <- run_model(sir, t = 0:100, n = 500)
plot(x)