Julia Implementation of Host SEIR + Vector SEI
Author: Carl A. B. Pearson @pearsonca
Date: 2018-10-02
This version considers multiple hosts, which introduces some indexing complications. This approach adopts the indexing scheme of:
- all of host $i$ compartments ($S_H$, $E_H$, etc), for $i \in 1\ldots N$
- vector compartments last
With this approach, we can re-use the solution for one host, with slight modifications:
H_comps = 4
V_comps = 3
# in sub functions, du / u are the particular relevant slices only
function F1H(du,u,p,t,I_V,N_H)
S_H, E_H, I_H, R_H = u
# host dynamics
host_infection = (p.β*S_H*I_V)/N_H
host_mortality = p.μ_H .* u # include S_H, so easier to remove mortality
host_births = sum(host_mortality)
host_progression = p.σ_H*E_H
recovery = p.λ*I_H
du[1] = -host_infection + host_births
du[2] = host_infection - host_progression
du[3] = host_progression - recovery
du[4] = recovery
du[1:end] -= host_mortality
end
# in sub functions, du / u are the particular relevant slices only
function FV(du,u,p,t,sum_β_I_H)
S_V, E_V, I_V = u
vec_infection = sum_β_I_H*S_V/p.N_H
vec_mortality = p.μ_V .* u # include S_V, so easier to remove mortality
vec_births = sum(vec_mortality)
vec_progression = p.σ_V*E_V
du[1] = -vec_infection + vec_births
du[2] = vec_infection - vec_progression
du[3] = vec_progression
du[1:end] -= vec_mortality
end
function F(du,u,p,t)
uvec = @view(u[(p.nHosts*H_comps+1):end]) # grab the vector compartments
S_V, E_V, I_V = uvec
sum_β_I_H = 0.0
for host in 0:(p.nHosts-1)
slice = (1:H_comps).+(H_comps*host)
F1H(@view(du[slice]), @view(u[slice]), p.host[host+1], t, I_V, p.vec.N_H)
# must use @view here, so that these arrays can be modified in F1H
sum_β_I_H += p.host[host+1].β * u[slice[3]] # this host's I compartment
end
FV(@view(du[(p.nHosts*4+1):end]), uvec, p.vec,t,sum_β_I_H)
end
F (generic function with 1 method)
First, state initial conditions. This code generates them randomly for convenience, though they could be assigned based on data, desired parameter space, or algorithmically as part of a fitting process:
nH = 5
srand(0)
S_Hs = ones(nH) .* 100.0
E_Hs = zeros(nH)
I_Hs = shuffle(vcat(zeros(nH-1),[1.0]))
R_Hs = zeros(nH)
host0 = reshape(hcat(S_Hs,E_Hs,I_Hs,R_Hs)', nH*4, 1)
vec0 = [10000.0, 0.0, 0.0]
u0 = vcat(host0, vec0)
23×1 Array{Float64,2}:
100.0
0.0
1.0
0.0
100.0
0.0
0.0
0.0
100.0
0.0
0.0
0.0
100.0
0.0
0.0
0.0
100.0
0.0
0.0
0.0
10000.0
0.0
0.0
Now, generate dynamic parameters. Again: this code generates them randomly for convenience, though they could be assigned based on data, desired parameter space, or algorithmically as part of a fitting process:
srand(1)
μs = 1 ./ (rand(nH) .* 360)
σs = 1 ./ (rand(nH) .* 6)
λs = 1 ./ (rand(nH) .* 28)
βs = rand(nH) ./ 10.0
using NamedTuples
# nb: in >= Julia v0.7, can eliminate this import
# and the @NT syntax
p = @NT(
nHosts = nH,
vec = @NT(μ_V=1/30, σ_V=1/7, N_H = sum(host0)),
host = [@NT(μ_H=μs[i], σ_H=σs[i], λ=λs[i], β=βs[i]) for i in 1:nH]
# just building up a random collection of params for demonstration
)
(nHosts = 5, vec = (μ_V = 0.03333333333333333, σ_V = 0.14285714285714285, N_H = 501.0), host = NamedTuples._NT_μ__H_σ__H_λ_β{Float64,Float64,Float64,Float64}[(μ_H = 0.0117686, σ_H = 0.790008, λ = 0.0642631, β = 0.0209472), (μ_H = 0.00801628, σ_H = 0.175085, λ = 0.0817059, β = 0.0251379), (μ_H = 0.00888301, σ_H = 0.166683, λ = 0.0840894, β = 0.00203749), (μ_H = 0.351205, σ_H = 0.662263, λ = 0.0461889, β = 0.0287702), (μ_H = 0.00568503, σ_H = 0.168919, λ = 0.127011, β = 0.0859512)])
Now these values can be used with the ODE solver:
using DifferentialEquations
using IterableTables, DataFrames
tspan = (0.0, 365.0)
prob = ODEProblem(F, u0, tspan, p)
sol = solve(prob,Tsit5(),reltol=1e-8,abstol=1e-8,saveat=linspace(0,365,365*10+1))
retcode: Success
Interpolation: 1st order linear
t: 3651-element Array{Float64,1}:
0.0
0.1
0.2
0.3
0.4
0.5
0.6
0.7
0.8
0.9
1.0
1.1
1.2
⋮
363.9
364.0
364.1
364.2
364.3
364.4
364.5
364.6
364.7
364.8
364.9
365.0
u: 3651-element Array{Array{Float64,2},1}:
[100.0; 0.0; … ; 0.0; 0.0]
[100.001; 4.05106e-8; … ; 0.041287; 0.000295821]
[100.002; 3.15487e-7; … ; 0.08154; 0.00117209]
[100.004; 1.03672e-6; … ; 0.120779; 0.00261227]
[100.005; 2.39309e-6; … ; 0.159026; 0.00460017]
[100.006; 4.55256e-6; … ; 0.196298; 0.00711996]
[100.007; 7.66377e-6; … ; 0.232616; 0.0101561]
[100.008; 1.18579e-5; … ; 0.267998; 0.0136934]
[100.009; 1.72497e-5; … ; 0.302464; 0.017717]
[100.011; 2.39395e-5; … ; 0.336032; 0.0222123]
[100.012; 3.20139e-5; … ; 0.36872; 0.0271651]
[100.013; 4.15473e-5; … ; 0.400545; 0.0325615]
[100.014; 5.26028e-5; … ; 0.431526; 0.0383877]
⋮
[45.2552; 0.790354; … ; 78.2356; 335.113]
[45.2574; 0.790394; … ; 78.2385; 335.114]
[45.2595; 0.790434; … ; 78.2413; 335.114]
[45.2617; 0.790474; … ; 78.2442; 335.115]
[45.2639; 0.790514; … ; 78.2471; 335.116]
[45.2661; 0.790553; … ; 78.25; 335.117]
[45.2682; 0.790593; … ; 78.2528; 335.117]
[45.2704; 0.790633; … ; 78.2557; 335.118]
[45.2725; 0.790673; … ; 78.2586; 335.119]
[45.2747; 0.790713; … ; 78.2615; 335.12]
[45.2768; 0.790753; … ; 78.2643; 335.121]
[45.279; 0.790792; … ; 78.2672; 335.122]
# rename!(df, Dict(:timestamp => :t,
# :value1 => :S_H, :value2 => :E_H, :value3 => :I_H, :value4 => :R_H,
# :value5 => :S_V, :value6 => :E_V, :value7 => :I_V
# ))
# mlt[:host] = contains.(string.(mlt[:variable]),"H"); # tag which entries are host vs vector
# df
df = DataFrame(sol)
mlt = melt(df,:timestamp) # convert results into long format for plotting
mlt[:index] = parse.(Int,replace.(string.(mlt[:variable]),r"[^\d]+"=>""))
mlt[:name] = [
"S_H1","E_H1","I_H1","R_H1",
"S_H2","E_H2","I_H2","R_H2",
"S_H3","E_H3","I_H3","R_H3",
"S_H4","E_H4","I_H4","R_H4",
"S_H5","E_H5","I_H5","R_H5",
"S_V","E_V","I_V"
][mlt[:index]]
mlt[:facet] = replace.(string.(mlt[:name]),r"\w+_"=>"")
mlt[:compartment] = replace.(string.(mlt[:name]),r"_\w+"=>"")
mlt
| variable | value | timestamp | index | name | facet | compartment | |
|---|---|---|---|---|---|---|---|
| 1 | value1 | 100.0 | 0.0 | 1 | S_H1 | H1 | S |
| 2 | value1 | 100.0011761246569 | 0.1 | 1 | S_H1 | H1 | S |
| 3 | value1 | 100.00235062049974 | 0.2 | 1 | S_H1 | H1 | S |
| 4 | value1 | 100.00352325035139 | 0.3 | 1 | S_H1 | H1 | S |
| 5 | value1 | 100.00469378414253 | 0.4 | 1 | S_H1 | H1 | S |
| 6 | value1 | 100.00586199875751 | 0.5 | 1 | S_H1 | H1 | S |
| 7 | value1 | 100.00702767789407 | 0.6 | 1 | S_H1 | H1 | S |
| 8 | value1 | 100.00819061190617 | 0.7 | 1 | S_H1 | H1 | S |
| 9 | value1 | 100.00935059768018 | 0.8 | 1 | S_H1 | H1 | S |
| 10 | value1 | 100.01050743847112 | 0.9 | 1 | S_H1 | H1 | S |
| 11 | value1 | 100.01166094378736 | 1.0 | 1 | S_H1 | H1 | S |
| 12 | value1 | 100.01281092925007 | 1.1 | 1 | S_H1 | H1 | S |
| 13 | value1 | 100.01395721643992 | 1.2 | 1 | S_H1 | H1 | S |
| 14 | value1 | 100.01509963280547 | 1.3 | 1 | S_H1 | H1 | S |
| 15 | value1 | 100.01623801150974 | 1.4 | 1 | S_H1 | H1 | S |
| 16 | value1 | 100.01737219128947 | 1.5 | 1 | S_H1 | H1 | S |
| 17 | value1 | 100.01850201637215 | 1.6 | 1 | S_H1 | H1 | S |
| 18 | value1 | 100.01962733634535 | 1.7 | 1 | S_H1 | H1 | S |
| 19 | value1 | 100.02074800598677 | 1.8 | 1 | S_H1 | H1 | S |
| 20 | value1 | 100.02186388520346 | 1.9 | 1 | S_H1 | H1 | S |
| 21 | value1 | 100.02297483892835 | 2.0 | 1 | S_H1 | H1 | S |
| 22 | value1 | 100.0240807369486 | 2.1 | 1 | S_H1 | H1 | S |
| 23 | value1 | 100.02518145380708 | 2.2 | 1 | S_H1 | H1 | S |
| 24 | value1 | 100.02627686872802 | 2.3 | 1 | S_H1 | H1 | S |
| 25 | value1 | 100.02736686552852 | 2.4 | 1 | S_H1 | H1 | S |
| 26 | value1 | 100.0284513324241 | 2.5 | 1 | S_H1 | H1 | S |
| 27 | value1 | 100.02953016196817 | 2.6 | 1 | S_H1 | H1 | S |
| 28 | value1 | 100.03060325097552 | 2.7 | 1 | S_H1 | H1 | S |
| 29 | value1 | 100.03167050044642 | 2.8 | 1 | S_H1 | H1 | S |
| 30 | value1 | 100.03273181537787 | 2.9 | 1 | S_H1 | H1 | S |
| ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ | ⋮ |
Now that we have a solution, we want to view what is happening in host vs mosquito population:
using RCall
# current version RCall supports better transfers, which would simplify this mess
# but requires Julia v >= 0.7
vals = mlt[:value]
tstamps = mlt[:timestamp]
fcts = mlt[:facet]
comps = mlt[:compartment]
@rput vals tstamps fcts comps
R"
library(ggplot2)
suppressPackageStartupMessages(library(data.table))
dt <- data.table(t=tstamps, y=vals, species=fcts, compartment=comps)
ggplot(dt) + aes(x=t, y=y, color=compartment) + facet_grid(species ~ ., scale = 'free_y') +
theme_minimal() +
geom_line()
"
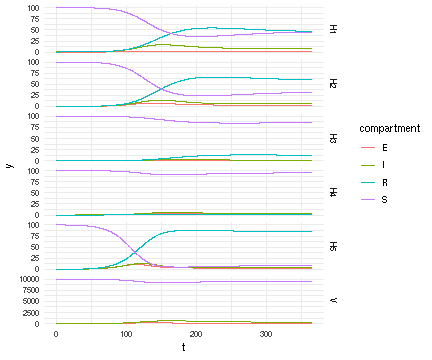
RCall.RObject{RCall.VecSxp}
